Case Study: Curing Disease with NumFOCUS Tools
NumFOCUS tools used to discover treatments and cures for disease

Photo credit: Maria Nemchuk (Broad Institute)
Dr. Anne Carpenter is an Institute Scientist and Imaging Platform Senior Director at the Broad Institute of Harvard and MIT. Her lab uses machine learning, especially deep learning, to capture information about genetic and chemical perturbations of cells in order to probe the causes and cures of disease.
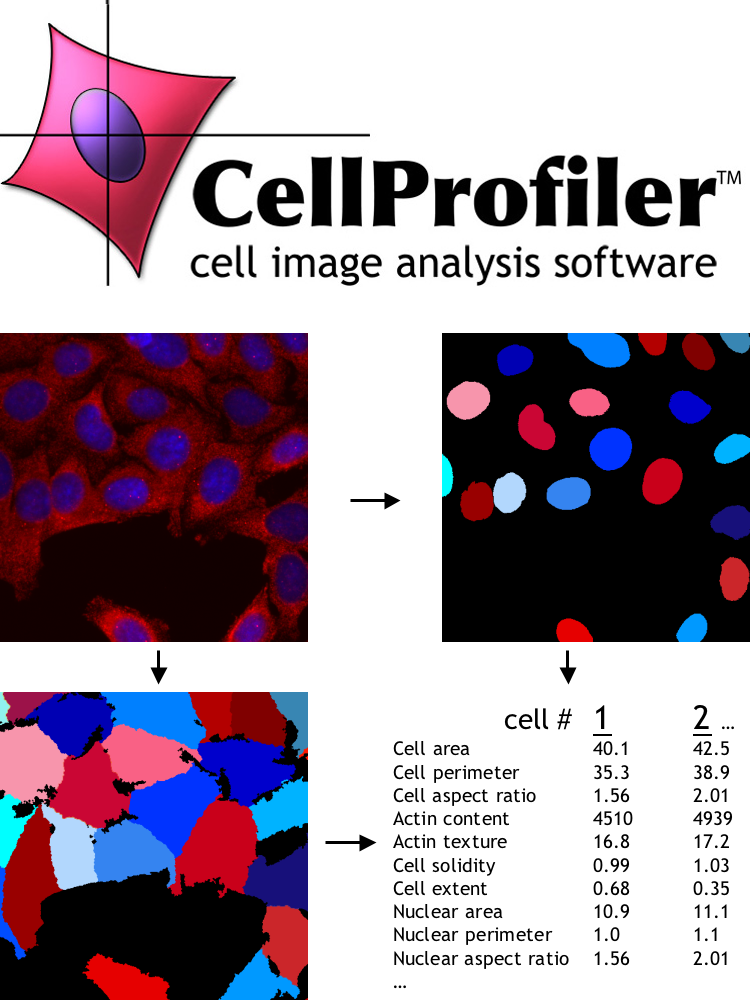
Carpenter co-created CellProfiler, now an open source Python tool that quantitatively analyzes and tracks the size and shape of cells. Because changes in cells are frequently associated with a variety of types of disease (cancer, for example), CellProfiler enjoys wide adoption by biologists conducting biomedical research. CellProfiler is built on top of NumFOCUS-supported open source projects, including NumPy, SciPy, Matplotlib, scikit-image and scikit-learn.
The creation story of CellProfiler is emblematic of the way in which scientific research software is developed, deployed, and eventually widely adopted—often somewhat by happy accident. It also illustrates the ongoing challenge of finding sustainable funding models for maintenance of critical research tools. NumFOCUS is engaged in tackling this challenge through our Sustainability Program, which convenes open source software project leaders for knowledge sharing and idea generation to identify and test potential solutions.
CellProfiler is built on top of NumFOCUS-supported open source projects, including NumPy, SciPy, Matplotlib, scikit-image and scikit-learn.
NumPy

SciPy
Matplotlib
Origins of a Scientific Software Project
When Carpenter started her postdoctoral fellowship in cell biology at the Whitehead Institute at MIT, her goal was to ultimately figure out how cell growth and cell size is regulated: an important question in understanding how cancer works. Carpenter quickly set out to find a software tool to help her measure those cells.
Unfortunately, in the early aughts there was neither commercial nor open source software available with the relevant capability for her research: to identify individual nuclei of fruit fly cells. By searching PubMed (a database of life sciences and biomedical research publications), Carpenter was able to find some computational papers that described an algorithm that would solve her problem. The algorithm did a good job of finding cells and measuring them, but the authors of the paper didn’t actually provide any code, much less a user-friendly tool that a biologist like herself could use. So, Carpenter did what most enterprising scientists do and decided to figure out how to build what she needed herself. But first, she’d have to learn how to code.
When Scientists Become Programmers
To start out, Carpenter hired a graduate student with programming experience to implement the algorithm described in the paper over the course of a weekend. Then Carpenter herself built out the user interface and the framework around that algorithm (in Matlab, which she taught herself). And as it turns out, she loved coding!
“This is something I was born for, and it’s too bad nobody told me, ‘Hey, you should be a software engineer,’ somewhere along the way,” Carpenter said. “So, better late than never, and in retrospect of course I’m thrilled that I switched fields because I bring a really unique perspective to developing software for biologists.”
In retrospect, there were hints that Carpenter was already pretty interested in technology. During her Ph.D. she worked on putting together an automated microscope that was necessary for a project, and towards the end of her doctoral program she started playing around with ImageJ, an open source image processing program. Carpenter’s biological interests were broad, and she already knew she was more interested in the technology than the biological questions in her research.
“I’m thrilled that I switched fields because I bring a really unique perspective to developing software for biologists.”

“For several years, I was the primary user of the software and the creator of the software, so I was able to think like a biologist and make an interface that was sensible for a biologist.” After a few months of building out features on CellProfiler, Carpenter realized that the research software she was creating could potentially be useful to a lot of people. “If you had told me, ‘Your job for the next 2 years is to build an enormous toolbox that thousands of biologists are going to use,’ I would have freaked out completely,” she said. “I just wrote it for my project and couldn’t imagine there was such a big need in the world that I, personally, would be able to fill—and that became pretty addictive. Once it became clear a lot of people needed the tool, I actually ended up abandoning my actual biological project” to focus working solely on the software.
“I just wrote it for my project and couldn’t imagine there was such a big need”
Prior to CellProfiler, biologists would look through their microscopes, maybe capture some digital images, and qualitatively describe what they saw. It was rare at the time for them to quantify what they saw, which is what CellProfiler is capable of doing—and why it was able to gain traction so quickly. Judging by published papers that cite the software, thousands of biologists around the world now use CellProfiler in their work.
Today, Carpenter finds herself in the somewhat unusual situation of being a cell biologist leading a group with no microscopes, no incubators, no pipettes: just computers. “What I love about my lab is that every day we hear about a different disease area that someone needs help on and a different biological area. So, I am a biologist; I love thinking about biological problems. It’s just I want to hear about two or three different ones every day as opposed to thinking about some magnificently narrow thing for the rest of my life. And so it really suits me well to be working on the technology that fuels a lot of different biology questions.”

The Shift to Open Source
In 2010, Carpenter was awarded her first National Institutes of Health (NIH) R01 grant, “Continued development of CellProfiler cell image analysis software.” That same year, she decided that CellProfiler needed to be decoupled from Matlab because of the issues it was causing. Whenever Matlab made an update, something catastrophic would happen with the CellProfiler code, which put them in a tough position. Furthermore, the expense of using proprietary software had become unsustainable for her research community: a lot of biologists were starting to do high-throughput computing and couldn’t afford a Matlab license for every node of their cluster.
She sent out a poll to determine what language they should rewrite the tool in, and the result came back roughly 50/50 between Java and Python. In fact, it was so close that her team had to make the language decision on their own. “If four people had voted the other way, it would be in Java right now,” she said. Based on the momentum they saw in the Python community and the upward direction it was trending, the people in her group had a slight preference for Python, so that’s what they chose. The rest is history, although Carpenter acknowledges that there would have been pros and cons to either choice. Even today, there are Java projects that they really want to tightly interoperate with (for example, ImageJ), which is challenging now that CellProfiler is written in Python.
the expense of using proprietary software had become unsustainable for her research community
Challenges of Scientific Software Maintenance: Growing a Contributor Community
To keep open source projects like CellProfiler functioning well, it helps to have a large-ish community of programmers who can contribute code to the project. One of the challenges of making open source software to serve researchers who aren’t programmers (like most biologists) is that it’s difficult to cultivate a community of contributors to maintain the code.
CellProfiler relies upon NumPy, SciPy, scikit-image, and scikit-learn (all NumFOCUS projects). Because those projects serve other computer scientists, it’s relatively easier for them to recruit a community of programmers to contribute code and fix bugs. Case in point: portions of code contributed to scikit-image and scikit-learn came originally from CellProfiler. “But when you’re making something that’s end-user facing, I think it’s a little bit more challenging,” according to Carpenter.

Researchers themselves are typically stretched thin already—Carpenter and her team don’t have the extra bandwidth to develop pathways to attract and onboard new contributors. Conversely, folks that do want to make code contributions are often more interested in building new features than maintaining the existing source code. Thus, for most of the lifespan of the software, CellProfiler has been maintained by just one (paid) programmer at a time. For a couple of years there were two, which allowed the project to become a little more open to outside contributions for a time.
And while one might be tempted to think that specialized knowledge in biology is a prerequisite for contributing to CellProfiler, Carpenter says that’s not the case at all. “You don’t have to have tremendous skill to get started being helpful in the open source community.” A codebase like CellProfiler that is oriented to biologists is a great place to get started as an open source contributor, “because everyone is just so thrilled to get the help!” Even new contributors can really make a difference to this type of project.
“You don’t have to have tremendous skill to get started being helpful in the open source community.”
When a button isn’t working, you don’t need the perspective of a biologist, says Carpenter. “The perspective of a biologist is necessary for writing really great documentation, for example, and it’s necessary for the overall design of the software, but I would say the vast majority of open issues right now have nothing to do with biology.” There are plenty of beginner-level issues available for new contributors to work on (and they are marked as such on GitHub).
Carpenter welcomes contributions from non-biologists because she doesn’t see a future in which most research scientists will also be programming their own research software, even if there has been a clear shift in the tech skills of the average biologist. “Everyone in every field is becoming a little more tech-savvy over time simply because that’s the world we live in. But I think it’s naive to say, ‘oh, well, everyone’s going to be at the command line in a decade or two.’” To Carpenter, that doesn’t necessarily represent a problem. “Time spent learning how to do things at the command line is time spent away from learning other things. And so yes, it’s awesome to have people who have both sets of skills, but I’m pretty passionate about creating software that serves both ends of the spectrum of the biology community.”
Challenges of Scientific Software Maintenance: Measuring Impact
What does work in CellProfiler’s favor is that the software’s impact is very compelling: it serves end-users who are working on discovering drugs and new mechanisms to treat disease. In contrast, the foundational nature of many NumFOCUS projects means that they are frequently multiple steps removed from their most impressive impacts.
Carpenter said she understands: “When you’re making the underlying libraries for things, it’s a little harder to point to the direct impact that it has. But I think anybody on the NumPy, SciPy, scikit-image teams can be really proud of—they are absolutely welcome to take credit for—all the things CellProfiler does, because we’re built on that foundation.”
“anybody on the NumPy, SciPy, scikit-image teams can be really proud of—they are absolutely welcome to take credit for—all the things CellProfiler does, because we’re built on that foundation.”
CellProfiler has been used to discover drugs that are on their way towards clinical trials, which takes a long time. There is one in clinical trials now, but that is because it was already a known drug and research suggested reusing an existing drug for a new type of cancer. Carpenter estimates that probably within a couple of years the first drugs based on using CellProfiler will come through. Her estimate is based on the work she knows that originates from academia, which links back to CellProfiler via citations in journal articles. It’s not clear, though, how her team would know if a pharmaceutical company had put something into clinical trials that used CellProfiler, because they have no means to track it. This is a common challenge when it comes to tracking the impacts of open source tools; the nature of the licensing and installation process means that it’s unusual for the creators to learn who is using their tools and to what ends.
One recent and innovative use of CellProfiler is actually using the software in the clinical trial. “They take cells from a patient, they use microscopy to image them and see how they respond to different potential candidate drug treatment, and then whichever cells respond the best—as measured by the software—that’s the drug that they choose to give the patient,” explained Carpenter. “So it’s kind of a personalized medicine approach, and the software is actually being used in that process, which is pretty exciting.”
Challenges of Scientific Software Maintenance: Funding
From 2003 to 2006, work on CellProfiler was funded through Carpenter’s postdoctoral fellowship. From 2007 to 2010, startup funds for her lab at the Broad Institute covered the cost of a software engineer. CellProfiler was funded by Carpenter’s NIH grant from 2010 to 2017, which she describes as “awesome.” And yet, the NIH has since discontinued its “Continued Maintenance and Development of Software” funding program. While there is a collection of other NIH funding programs, as a rule (whether written or unwritten) they all emphasize new feature development rather than maintenance and support.
“Financing the project is my biggest concern,” Carpenter said. “It’s insane to me that it only costs one software engineer to keep this project for thousands of people alive. That’s not the biggest investment in the world, but it’s really challenging for funding agencies like the National Institutes of Health—nobody wants to fund maintenance.”
“It’s insane to me that it only costs one software engineer to keep this project for thousands of people alive.”


Funding maintenance is important because when a new operating system comes out or a new dependency changes, the software must be updated to remain functional. Unfortunately, the major funding agencies only support grants for new projects and have been especially reluctant to fund technical support for existing projects. “That’s our biggest challenge, I would say, is keeping the project funded. Making it self-sustaining, short of ad-ware—I don’t see how it’s possible without some amount of funding.” Carpenter has tried to piece together bits and pieces where she can.
Just recently, the Chan Zuckerberg Initiative noticed this problem and decided to do something about it, in the form of financially supporting a group of three Software Fellows working on the most critical open-source bioimaging software packages. The cohort includes Allen Goodman in the Carpenter lab to support CellProfiler, as well as Juan Nunez-Iglesias supporting scikit-image and Curtis Rueden supporting FIJI/ImageJ. “When CZI scientists told me they were interested in funding open bioimaging software, I literally had to fight back tears… it was so rewarding to finally have someone recognize the importance of financially supporting this kind of work.” Like many open source project leaders, she and her team are still attempting to figure out a long-term solution to the challenge of maintaining the CellProfiler codebase.
Carpenter emphasizes that computational infrastructure work on projects like NumPy and SciPy requires philanthropy through an organization like NumFOCUS, because the support won’t happen otherwise. “It’s so catalytic, it’s so multiplicative. The amount of money that goes into an infrastructural tool and maintenance of a tool is so much better than adding some feature to something somewhere.”
“It doesn’t take a huge dollar amount, or a huge amount of bug-fixing, to support software in a way that is meaningful.”
According to Carpenter, people might be really shocked at how much of a difference their money and time can make in open source software. “It doesn’t take a huge dollar amount, or a huge amount of bug-fixing, to support software in a way that is meaningful.”



































